Sample queries of DEDAL on Essentia Proteomica server
The saposin/"swaposin" example
Query
This example presents a typical case of a circular permutation. The circular permutation between saposin and saposin-like “swaposin” domains is one of the very first known. It was discovered using sequence analysis, and verified when the crystal structures became available. NK-lysin (SCOP domain
d1nkla_) comprises five α-helices of the “folded leaf” architecture. The “swaposin” domain (
d1qdma1) of aspartic proteinase prophytepsin has the same architecture, but the helices are in a different order. Most of the structure comparison methods attempt to align the helices in agreement with their order along the sequence, which results in a visually poor superposition. They also fail to correctly align the cysteine residues forming the disulfide bonds. DEDAL correctly handles these tasks.
The screenshot below demonstrates a query for comparing these structures.
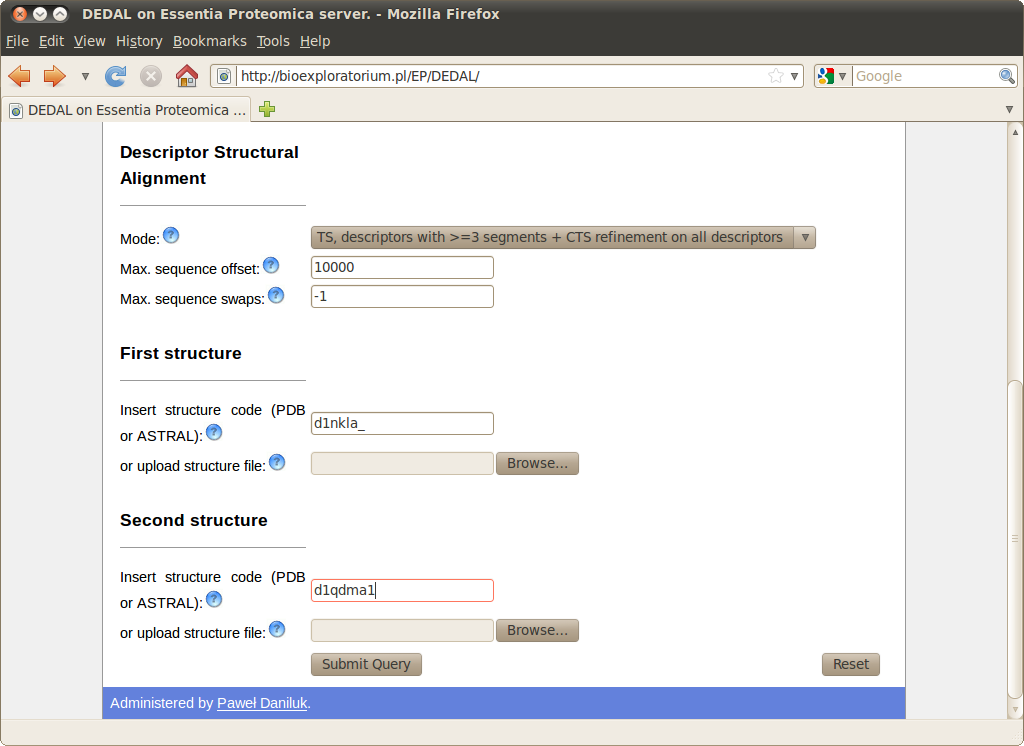
Computation
After successfully submitting the query user receives a message with a link to the computation results.
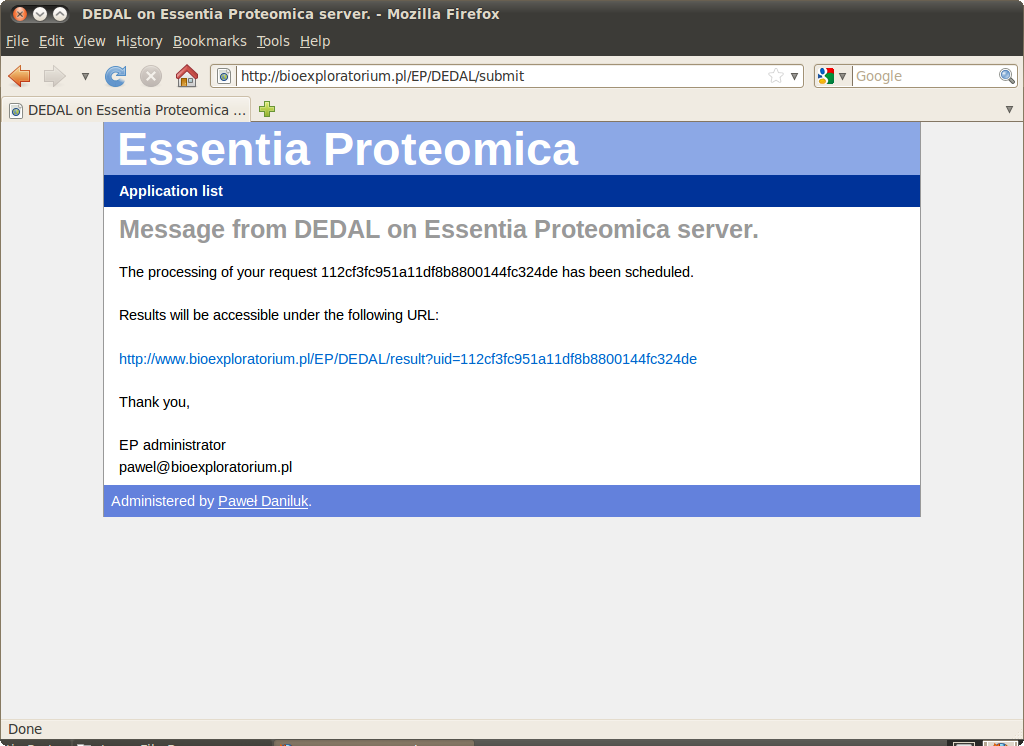
Although it may take several minutes to queue and execute a job the link is active and redirects to an automatically refreshing page informing, that the query is being processed.
Results
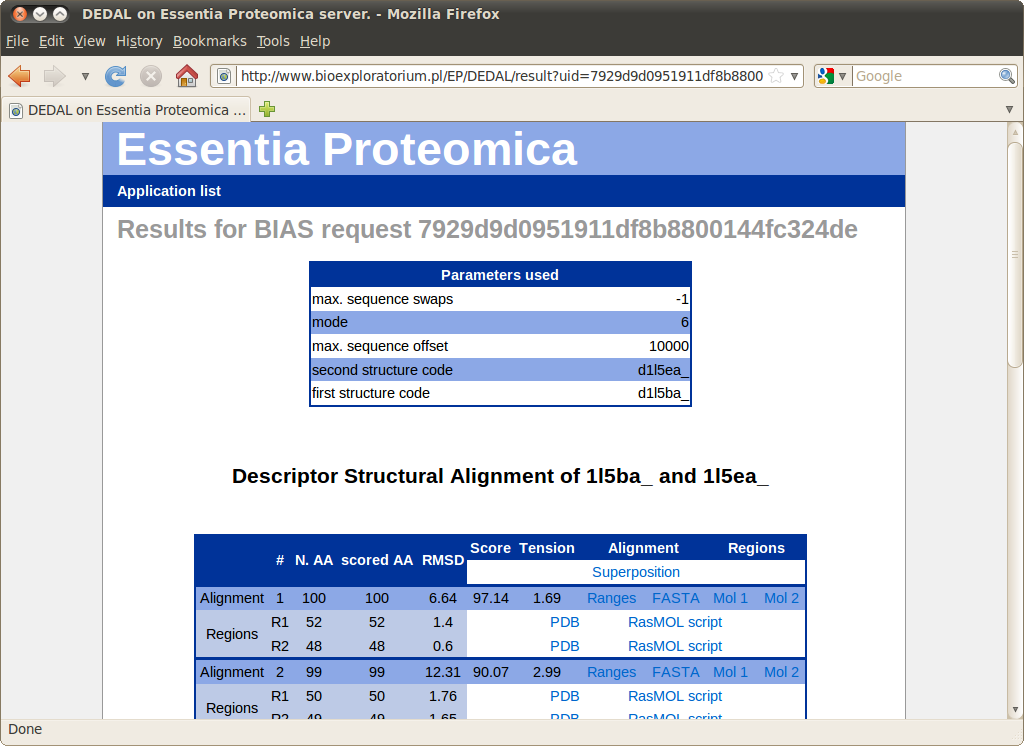
At most 5 highest scoring alignments are presented. Each alignment comprises one or more superimposable regions. The table shows the size, RMSD and "tension" for each region as well as the whole alignment.
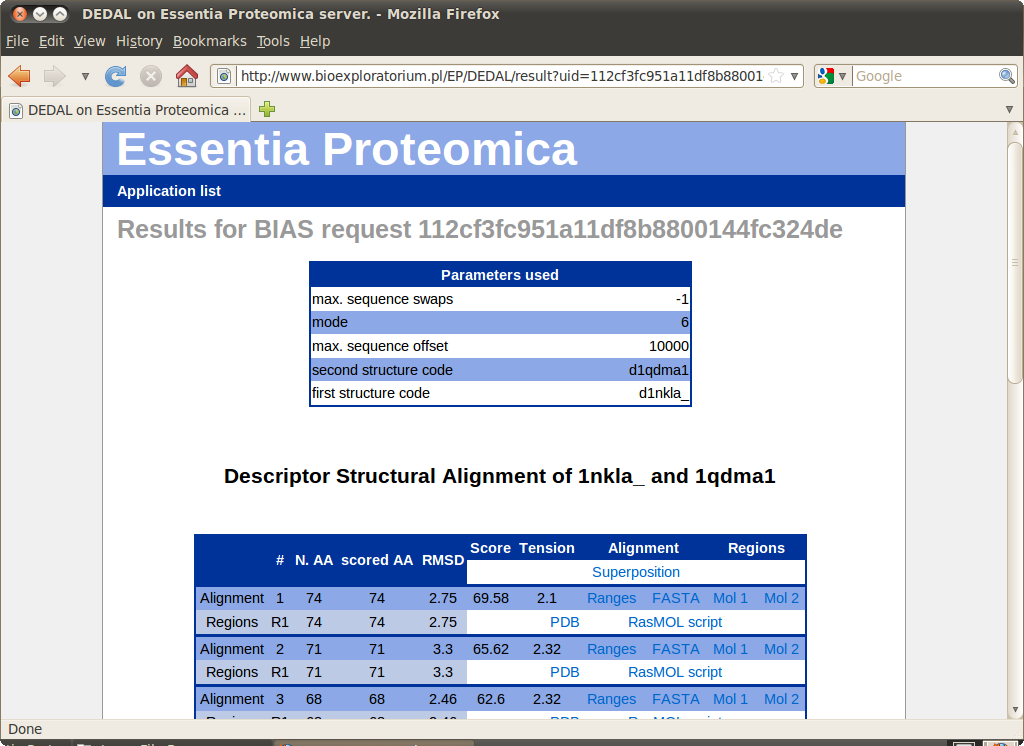
The following files are available for download for each alignment:
- Text file with aligned ranges
- FASTA file with sequence alignment
- PDB file with superpositions for each region
- RasMOL scripts with colored superpositions for each region
- RasMOL scripts with regions highlighted on each structure
Superpositions with highlighted aligned ranges can be viewed in RasMOL.
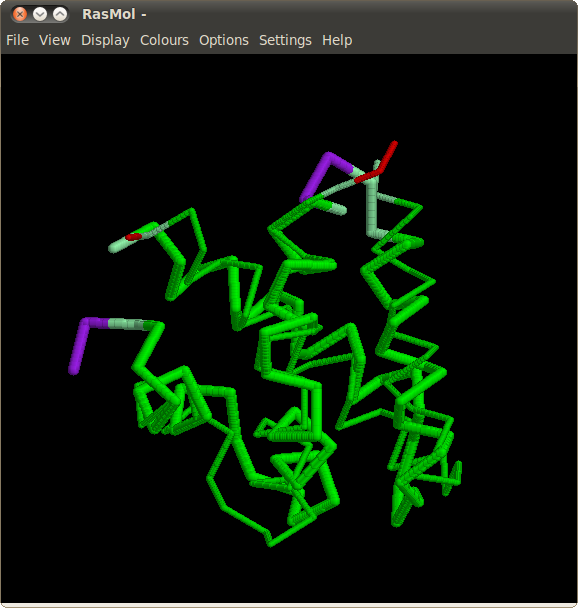
Cyanovirin-N
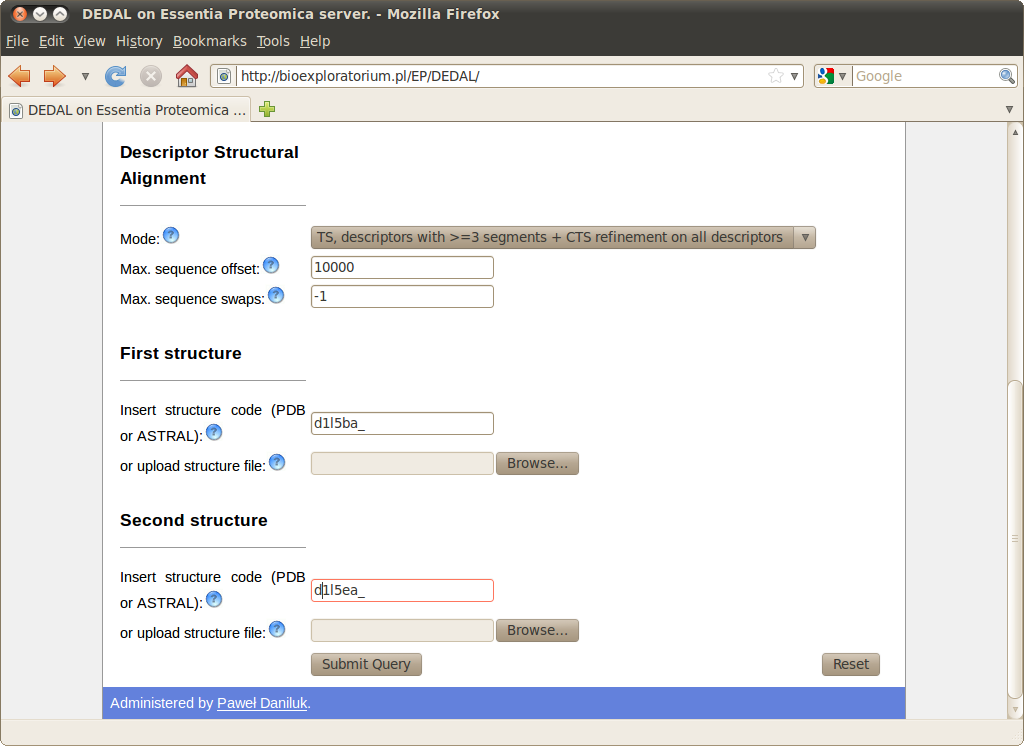
Query
Cyanovirin-N is a potent HIV-inactivating protein, present in both monomeric and domain-swapped dimeric forms. Although the monomeric form is predominant in solution, and was determined first, the metastable dimeric form is also present. The dimeric form is stabilized in the crystalline state and eventually its structure was also obtained by NMR. For the dimeric form, it can be observed that the X-ray (SCOP domain
d1l5ba_) and NMR (
d1l5ea_) structures have slightly different arrangement of subdomains, and that the local conformations of all residues except for the hinge region (PRO51-ASN53) are identical. Nevertheless the similarity between the two structures cannot be easily analyzed by using the rigid-body techniques.
Results
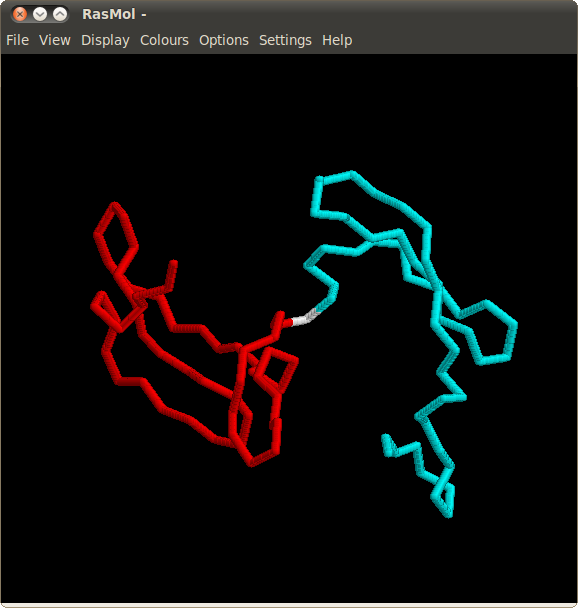
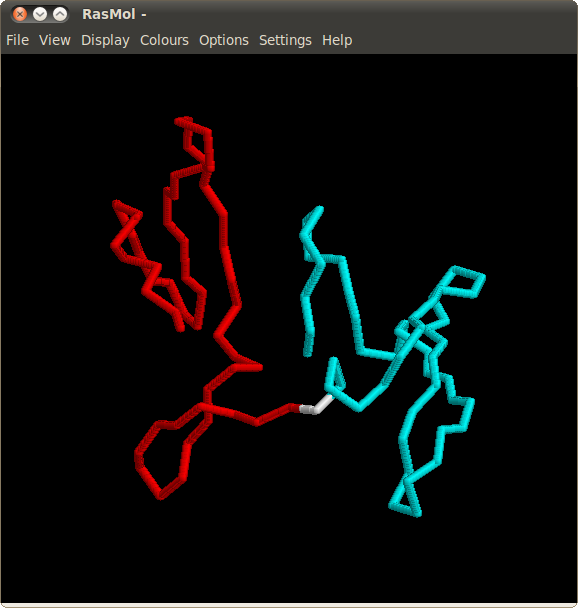
Alignments obtained for variants of Cyanovirin-N comprise two independent regions connected by a "hinge". The spatial relationship between regions can be easily visualized in RasMOL by opening links "Mol 1" and "Mol 2" in the "Regions" column.

 Although it may take several minutes to queue and execute a job the link is active and redirects to an automatically refreshing page informing, that the query is being processed.
Although it may take several minutes to queue and execute a job the link is active and redirects to an automatically refreshing page informing, that the query is being processed.

 The following files are available for download for each alignment:
The following files are available for download for each alignment:


 Alignments obtained for variants of Cyanovirin-N comprise two independent regions connected by a "hinge". The spatial relationship between regions can be easily visualized in RasMOL by opening links "Mol 1" and "Mol 2" in the "Regions" column.
Alignments obtained for variants of Cyanovirin-N comprise two independent regions connected by a "hinge". The spatial relationship between regions can be easily visualized in RasMOL by opening links "Mol 1" and "Mol 2" in the "Regions" column.






